Help
About YQFC
Usage of YQFC
About YQFC
Motivation of YQFC
Nowadays omics technologies have been routinely used in the biological research. From the omics data, researchers can easily get two gene lists (e.g. stress-induced genes vs. stress-repressed genes) related to their biological question. The next step is to apply enrichment analysis tools to identify distinct functional/regulatory features between these two gene lists for further investigation. Although various enrichment analysis tools are already available, two challenges remain to be addressed.
- Most existing tools are designed to analyze one gene list, so they cannot directly compare two gene lists.
- Almost all existing tools focus on identifying the enriched qualitative features (e.g. GO terms, pathways, domains, etc.).
Many quantitative features (e.g. # of mRNA isoforms of a gene, mRNA half-life, protein half-life, transcriptional plasticity, translational efficiency, etc.) are available in the yeast, but no existing tools provide analyses on these quantitative features. To address these two challenges, here we develop YQFC (Yeast Quantitative Features Comparator) to directly compare quantitative features between two yeast gene lists.
What is YQFC?
YQFC (Yeast Quantitative Features Comparator) is a webtool to directly compare quantitative features between two yeast gene lists. We comprehensively collected and processed 85 quantitative features from the yeast literature and yeast databases. For each quantitative feature, YQFC provides three statistical tests (t-test, U test, and KS test) to test whether this quantitative feature is statistically different between the two input yeast gene lists. The distinct quantitative features identified by YQFC may help researchers to study the underlying molecular mechanisms that differentiate the two input yeast gene lists. We believe that YQFC is a useful tool to expedite the biological research that uses the omics technologies.
Construction of YQFC
Collection of 85 quantitative features from seven yeast publications and six yeast databases
We comprehensive collected and processed 85 quantitative features from seven yeast publications (Holstege 1998, Nagalakshmi 2008, Lin 2010, Pelechano 2013, Christiano 2014, Csárdi 2015, Ho 2018) and six yeast databases (YeastMine, YeastNet, YAGM, SGD, BioGRID, SPELL).
We classified these 85 quantitative features into four categories: 12 gene features, 4 mRNA features, 52 protein features, and 17 network features.
| 12 Gene Features | CDS length, 5’UTR length, 3’UTR length, # of publications, # of GO terms, # of GO slim terms, # of pathways, # of mutant phenotypes, # of mRNA isoforms, # of transcriptional regulators, # of fungal homologs, # of non-fungal and S. cerevisiae homologs |
| 4 mRNA Features | mRNA level (3 datasets), mRNA half-life, transcriptional plasticity , translational efficiency (5 datasets) |
| 52 Protein Features | # of domains, # of PTMs, amino acid composition (20 kinds), atomic composition (5 kinds), protein half-life, protein abundance (in the normal growth condition, 23 datasets), protein abundance (in various stress conditions, 11 kinds), extinction coefficient at 280nm (2 kinds) , protein physical details (protein length, molecular weight, Isoelectric point , aliphatic index , instability index ), coding region translation calculation (codon bias , codon adaptation index , frequency of optimal codons , hydropathicity of protein , aromaticity score ), |
| 17 Network Features | # of interactors in the [GI , PI (3 datasets) , CC , CX , DC , GN , GT , PG , TS , EPA , FAA , GIA , LEA , MPA , PIA , TFBA , TFRA ] network |
The detailed source information of each individual feature is given in Supplementary Table 1.
Three statistical tests (t-test, U test, and KS test) used to compare a quantitative feature between two yeast gene lists
For each quantitative feature, YQFC provides three statistical tests (t-test, U test, and KS test) to test whether this quantitative feature is statistically different between the two given yeast gene lists (denoted as L1 and L2). For example, assume that a user wants to compare the 5’UTR length between L1 and L2. YQFC provides the following three statistical tests to do this task.
- The t-test (or called Student’s t-test) is used to determine if the mean 5’UTR length of the genes in L1 is statistically longer/shorter than the mean 5’UTR length of the genes in L2. Note that the t-test is a parametric test based on the assumption that the observed data come from normal distributions.
- The U test (or called Mann-Whitney U test) is used to determine if the median 5’UTR length of the genes in L1 is statistically longer/shorter than the median 5’UTR length of the genes in L2 under the location shift assumption. Note that the U test is a non-parametric test which does not assume anything about the distribution of the underlying populations.
- The KS test (or called Kolmogorov-Smirnov test) is used to determine if the cumulative distribution of the 5’UTR length of the genes in L1 is statistically larger/smaller than the cumulative distribution of the 5’UTR length of the genes in L2. Note that the KS test is a non-parametric test which does not assume anything about the distribution of the underlying populations.
Implementation of the web interface of YQFC
The following figure illustrates the configuration of YQFC.
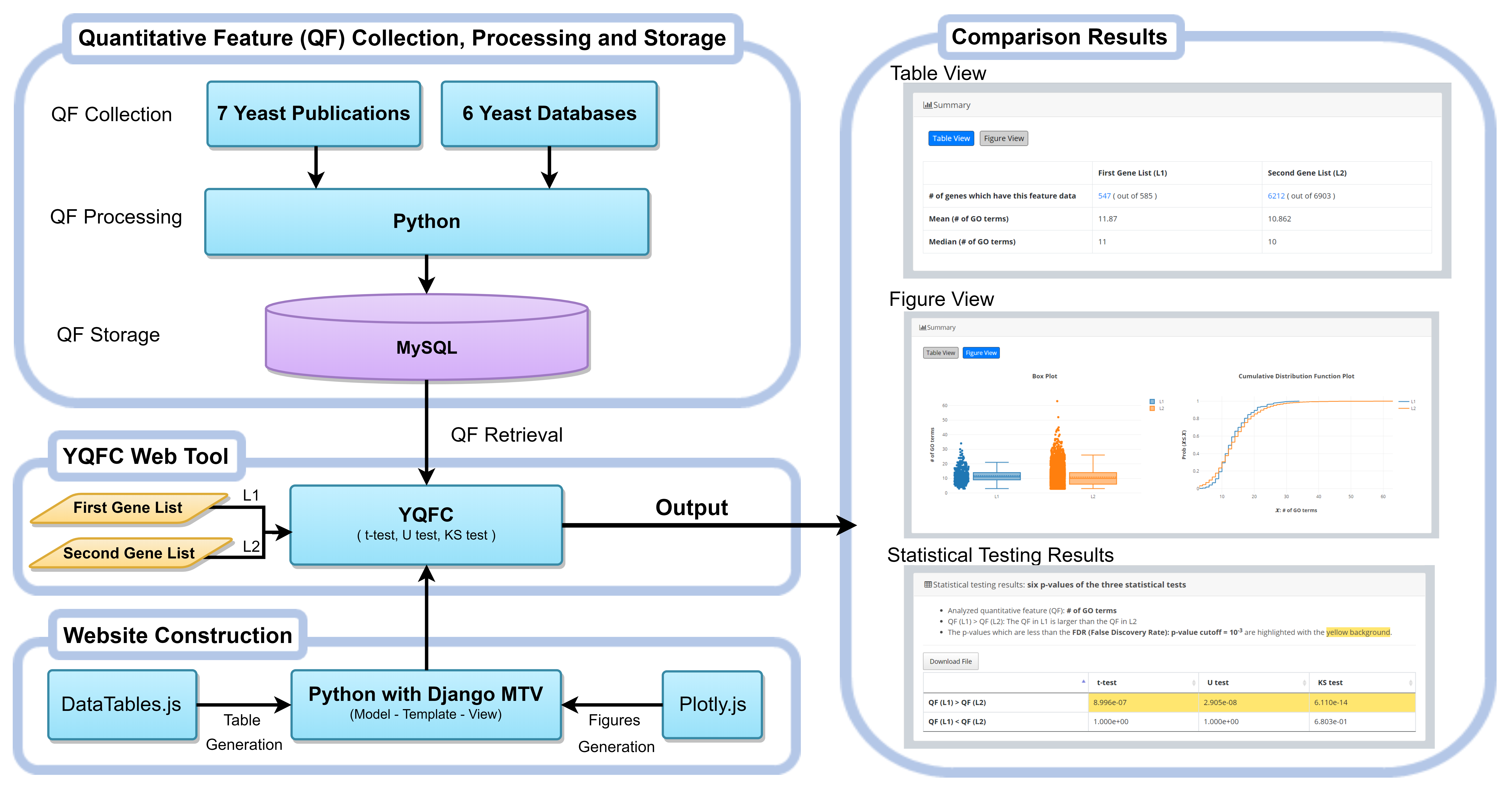
The web interface of YQFC was developed in Python using the Django MTV framework. The 85 processed quantitative feature data were deposited in MySQL. All tables, box plots, and cumulative distribution function (CDF) plots were produced by the JavaSscript and feature-rich JavaScript libraries (jQuery, DataTables and Plotly.js) to visualize data on the webpage. Except for the main website (http://cosbi2.ee.ncku.edu.tw/YQFC/), we also have two backup sites (http://cosbi4.ee.ncku.edu.tw/YQFC/) and (http://cosbi6.ee.ncku.edu.tw/YQFC/).
Usage of YQFC
The usage of YQFC
YQFC is a web tool for identifying the distinct quantitative features between two input yeast gene lists.
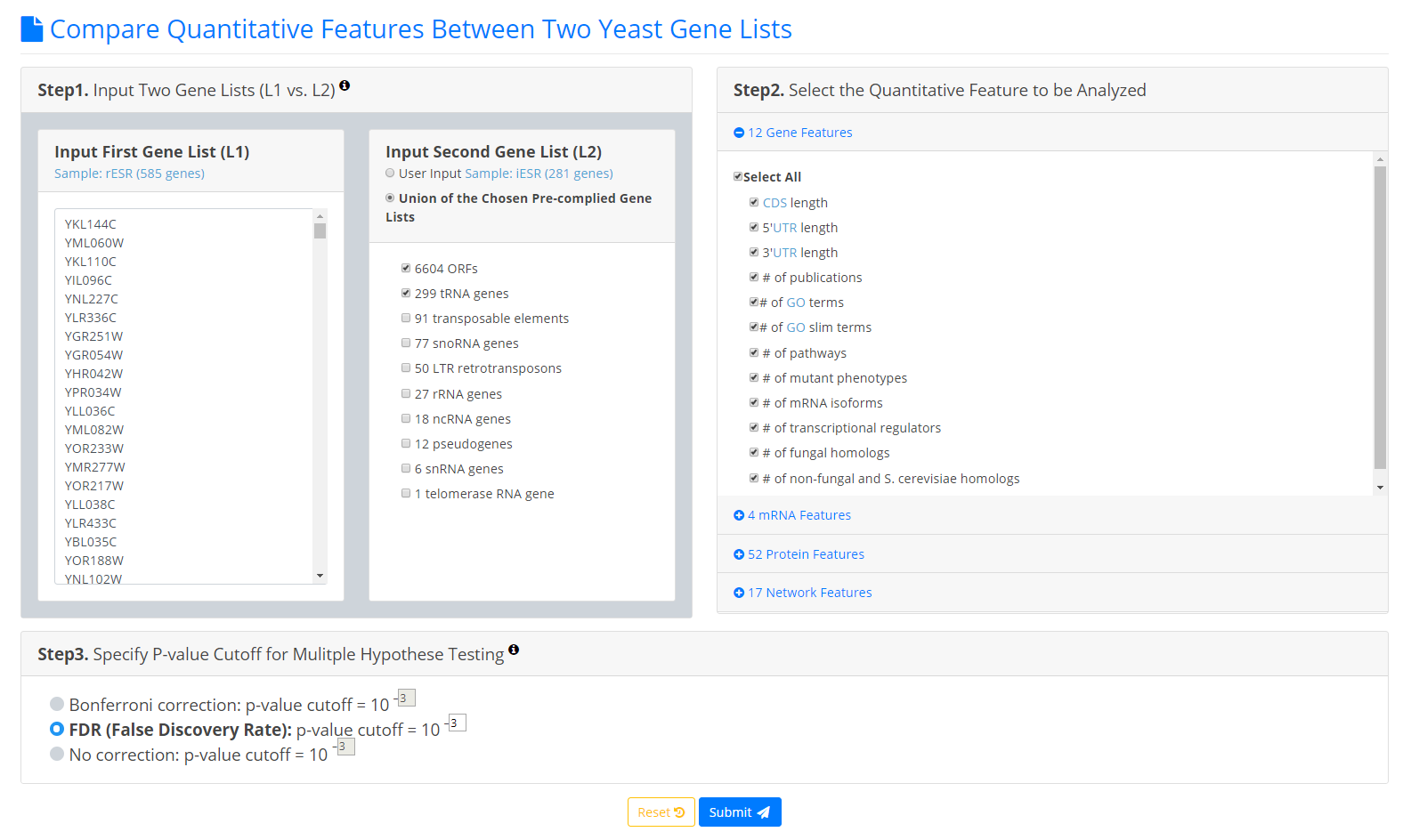
To use YQFC, users have to go through a three-step process.
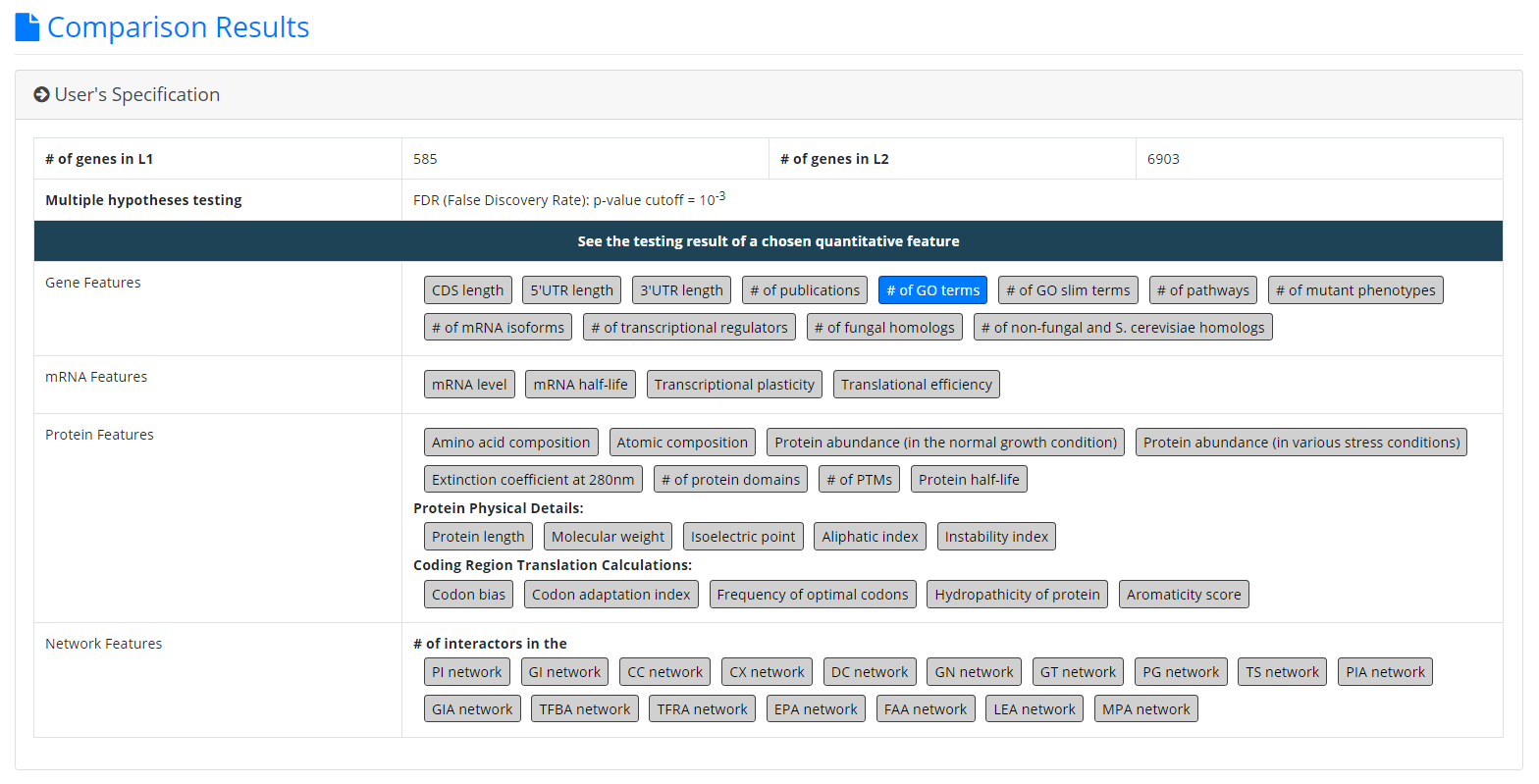
- Users need to input two gene lists to be compared. Standard names, systematic names, or aliases are all acceptable. If users only have one input gene list, they can use our pre-complied gene lists (e.g. 6604 ORF genes, 299 tRNA genes, 27 rRNA genes, etc.) to serve as the second input gene list.
- Users need to select the quantitative features to be analyzed.
- Since YQFC tests many quantitative features (i.e. multiple hypotheses testing), users have to select a statistical method (Bonferroni correction or FDR) for multiple hypotheses correction and set the p-value threshold. The p-value threshold determines the statistical significance of how different of a quantitative feature is between the two input gene lists. The more stringent the p-value threshold, the higher the statistical significance of the identified distinct quantitative feature.
After submission, YQFC will perform three statistical tests (t-test, U test, and KS test) to test whether a selected quantitative feature is statistically different between the two input gene lists (denoted as L1 and L2). This process will go through all the selected quantitative features. Once the analysis process is complete, YQFC will return the results with two parts.
- The first part contains the information of the user’s settings (i.e. the number of genes in L1, the number of genes in L2, the selected quantitative features, the selected multiple hypotheses correction method, and the selected p-value threshold).
- The second part contains the result of each selected quantitative feature shown as two sections: (i) Summary and (ii) Statistical testing results.
-
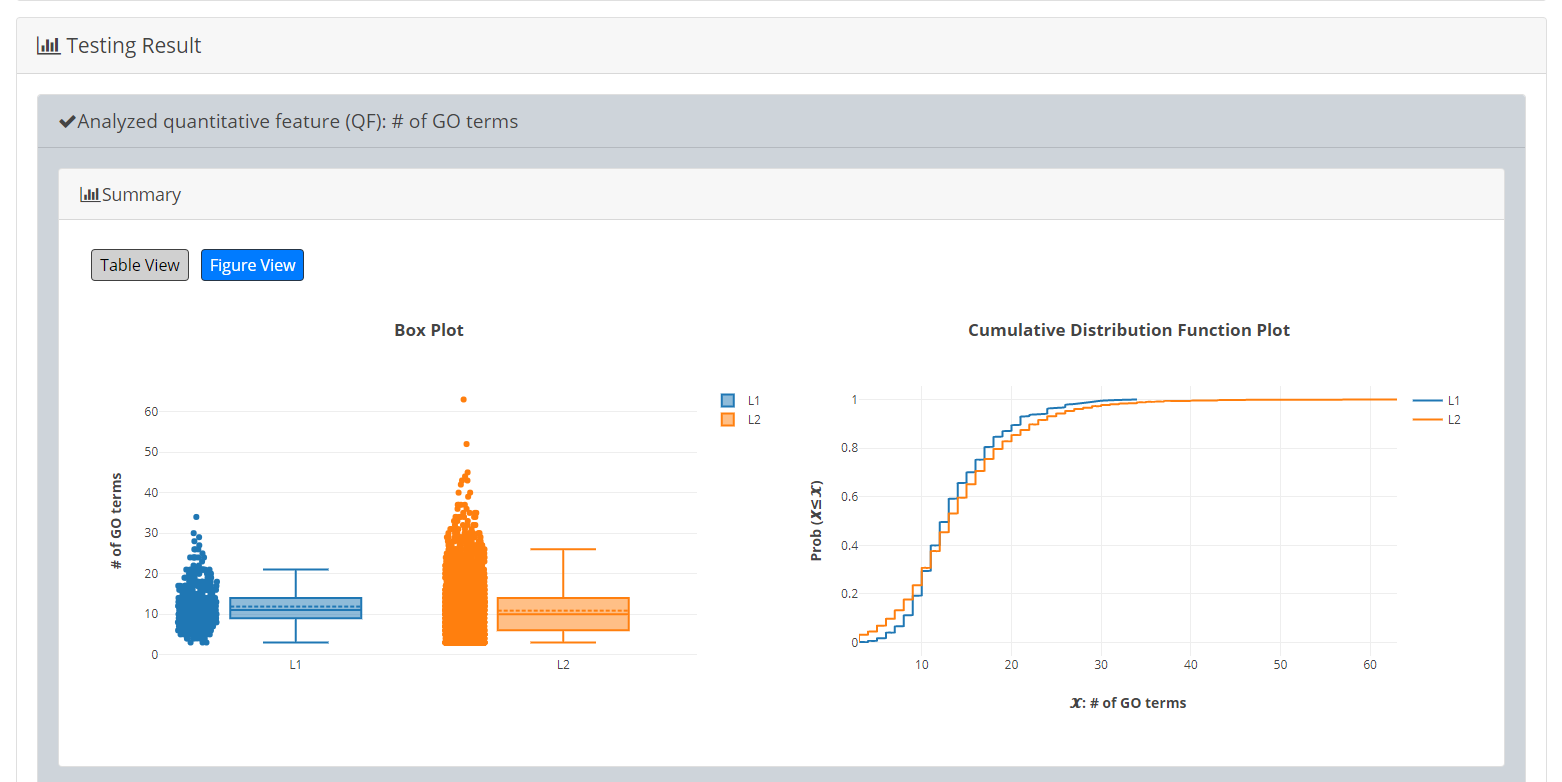
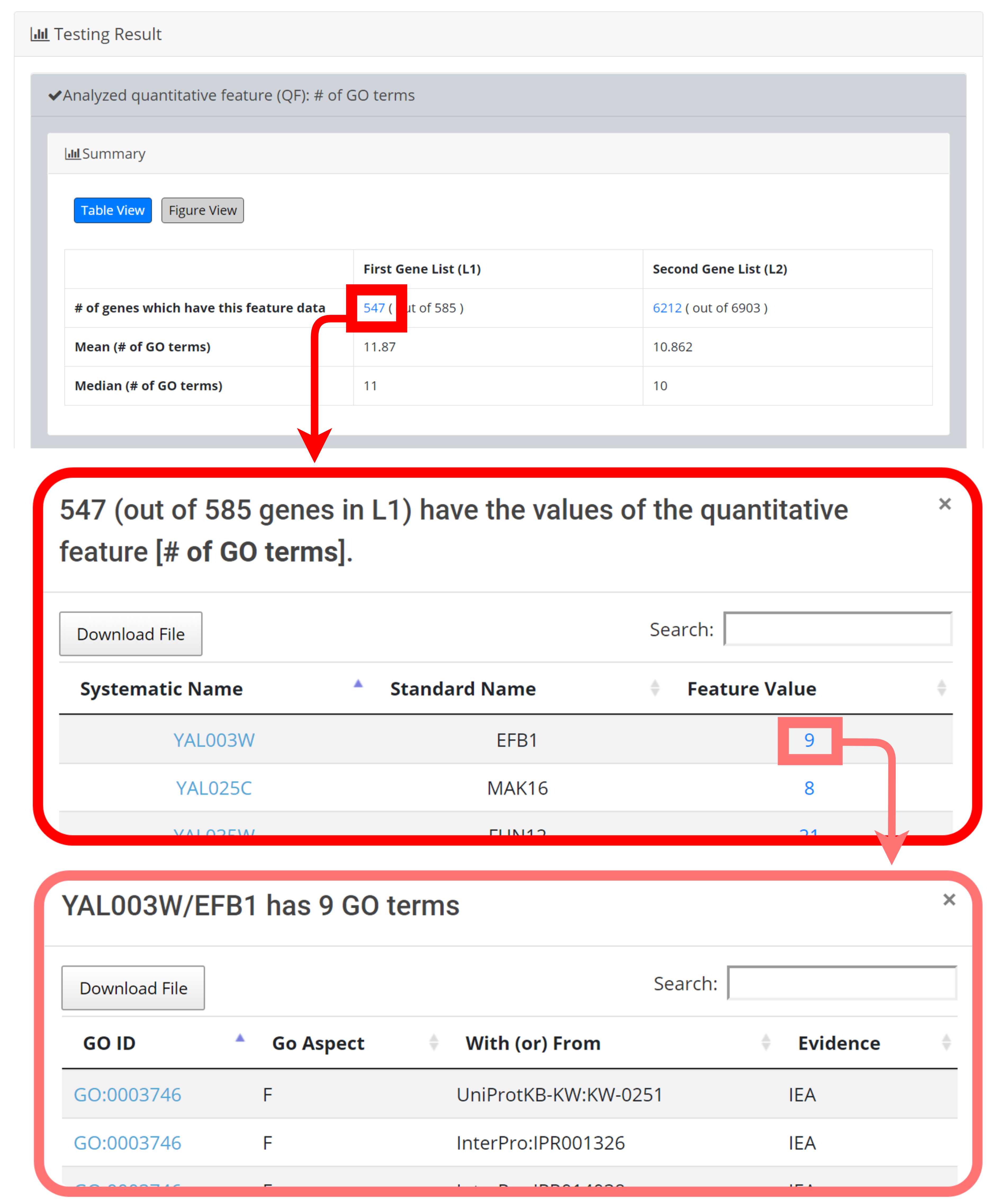
[Table View] provides a table containing two kinds of information. First, the numbers of genes (in L1 and L2, respectively) which have the feature values are given. If users click on the number, they will see the names and the feature values of these genes. By clicking on a feature value, users will see the original sources of the feature value. Second, the mean and median feature values of the genes (having feature values) in L1 and L2, respectively, are given.
- [Figure View] provides two kinds of plots for visualization. Box plots are used to display variation in the feature values in L1 and L2, respectively. Cumulative distribution function (CDF) plots are used to show the probability that the feature value 𝙓 is less than or equal to a specific value 𝓧 (i.e. Prob(𝙓≤𝓧)).
-
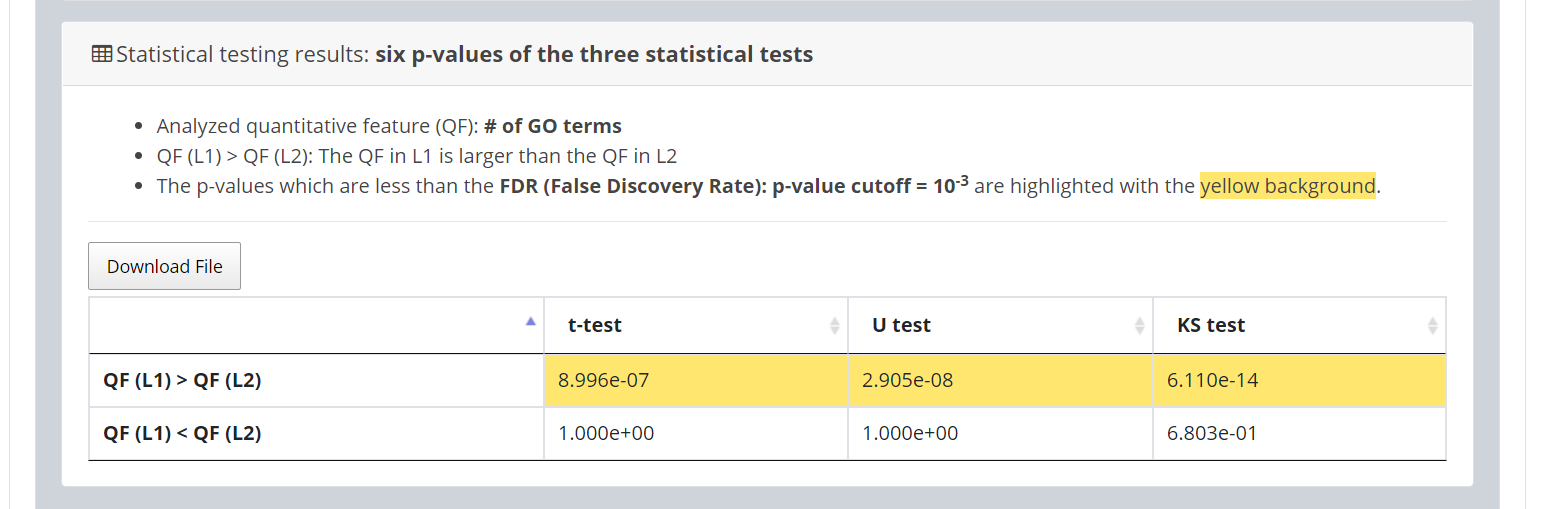
In the “Statistical testing results” section, users can see a table with six p-values. Three p-values (calculated by t-test, U test, and KS test) represent the statistical significance of claiming the quantitative feature in L1 is larger than that in L2 (denoted as QF(L1 )>QF(L2 )). The other three p-values (calculated by t-test, U test, and KS test) represent the statistical significance of claiming the quantitative feature in L1 is smaller than that in L2 (denoted as QF(L1)
< QF(L2)). To draw the users’ attention, the p-values which are less than the p-value threshold are highlighted with the yellow background.
In the “Summary” section, users can choose [Table View] or [Figure View].
- The second part contains the result of each selected quantitative feature shown as two sections: (i) Summary and (ii) Statistical testing results.